Working with ASE¶
The Atomic Simulation Environment (ASE) is a set of tools and Python modules for setting up, manipulating, running, visualizing and analyzing atomistic simulations.
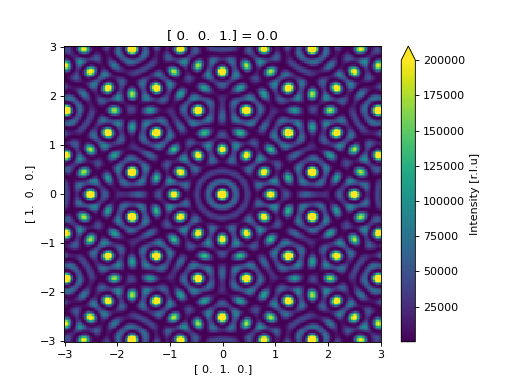
Using the ase.build.nanotube() as an example of using
calculating the scattering directly from an ase.Atoms object.
>>> from javelin.fourier import Fourier # doctest: +SKIP
>>> from ase.build import nanotube # doctest: +SKIP
>>> cnt = nanotube(6, 0, length=4) # doctest: +SKIP
>>> print(cnt) # doctest: +SKIP
Atoms(symbols='C96', pbc=[False, False, True], cell=[0.0, 0.0, 17.04])
>>> cnt_four = Fourier() # doctest: +SKIP
>>> cnt_four.grid.bins = 201, 201 # doctest: +SKIP
>>> cnt_four.grid.r1 = -3, 3 # doctest: +SKIP
>>> cnt_four.grid.r2 = -3, 3 # doctest: +SKIP
>>> results = cnt_four.calc(cnt) # doctest: +SKIP
Working on atom number 6 Total atoms: 96
>>> results.plot(vmax=2e5) # doctest: +SKIP
<matplotlib.collections.QuadMesh object at ...>
(Source code, png, hires.png, pdf)
Convert structure from ASE to javelin¶
A ase.Atoms can be converted to
javelin.structure.Structure simply by initializing the
javelin structure from the ASE atoms.
>>> print(cnt) # doctest: +SKIP
Atoms(symbols='C96', pbc=[False, False, True], cell=[0.0, 0.0, 17.04])
>>> type(cnt) # doctest: +SKIP
<class 'ase.atoms.Atoms'>
>>> from javelin.structure import Structure # doctest: +SKIP
>>> javelin_cnt = Structure(cnt) # doctest: +SKIP
>>> print(javelin_cnt) # doctest: +SKIP
Structure(C96, a=1, b=1, c=17.04, alpha=90.0, beta=90.0, gamma=90.0)
>>> type(javelin_cnt) # doctest: +SKIP
<class 'javelin.structure.Structure'>
Convert structure from javelin to ASE¶
To convert javelin.structure.Structure to ase.Atoms
you can use javelin.structure.Structure.to_ase().
>>> from javelin.io import read_stru # doctest: +SKIP
>>> pzn = read_stru('../../tests/data/pzn.stru') # doctest: +SKIP
Found a = 4.06, b = 4.06, c = 4.06, alpha = 90.0, beta = 90.0, gamma = 90.0
Read in these atoms:
O 9
Pb 3
Nb 2
Zn 1
Name: symbol, dtype: int64
>>> print(pzn) # doctest: +SKIP
Structure(O9Pb3Nb2Zn1, a=4.06, b=4.06, c=4.06, alpha=90.0, beta=90.0, gamma=90.0)
>>> type(pzn) # doctest: +SKIP
<class 'javelin.structure.Structure'>
>>> pzn_ase = pzn.to_ase() # doctest: +SKIP
>>> print(pzn_ase) # doctest: +SKIP
Atoms(symbols='PbNbO3PbZnO3PbNbO3', pbc=False, cell=[4.06, 4.06, 4.06])
>>> type(pzn_ase) # doctest: +SKIP
<class 'ase.atoms.Atoms'>
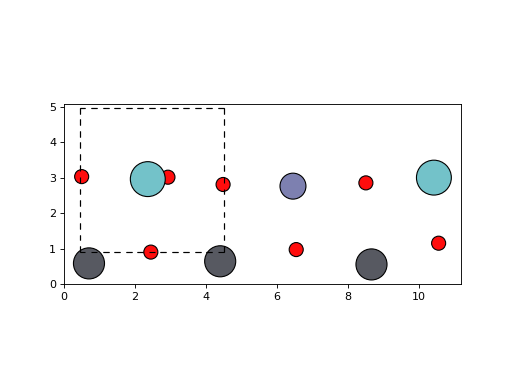
Visualization with ASE¶
The javelin.structure.Structure has enough api compatibility
with ase.Atoms that it can be used with some of ASE’s
visulization tools.
An example using ASE’s matplotlib interface.
>>> from javelin.io import read_stru # doctest: +SKIP
>>> from ase.visualize.plot import plot_atoms # doctest: +SKIP
>>> pzn = read_stru('../../tests/data/pzn.stru') # doctest: +SKIP
>>> print(pzn) # doctest: +SKIP
Structure(O9Pb3Nb2Zn1, a=4.06, b=4.06, c=4.06, alpha=90.0, beta=90.0, gamma=90.0)
>>> plot_atoms(pzn, radii=0.3) # doctest: +SKIP
<matplotlib.axes._subplots.AxesSubplot object at ...>
(Source code, png, hires.png, pdf)
To use all of ASE’s visualization tools, such as ase.gui, VMD,
Avogadro, or ParaView, first Convert structure from javelin to
ASE.
>>> from ase.visualize import view # doctest: +SKIP
>>> pzn_ase = pzn.to_ase() # doctest: +SKIP
>>> view(pzn_ase) # doctest: +SKIP
>>> view(pzn_ase, viewer='vmd') # doctest: +SKIP
>>> view(pzn_ase, viewer='avogadro') # doctest: +SKIP
>>> view(pzn_ase, viewer='paraview') # doctest: +SKIP
File IO¶
ase.io has extensive support for file-formats that can be
utilized by javelin. For example reading in ‘.cif’ files using
ase.io.read()
>>> from javelin.structure import Structure # doctest: +SKIP
>>> from ase.io import read # doctest: +SKIP
>>> graphite = Structure(read('tests/data/graphite.cif')) # doctest: +SKIP
>>> print(graphite) # doctest: +SKIP
Structure(C4, a=2.456, b=2.456, c=6.696, alpha=90.0, beta=90.0, gamma=119.99999999999999)
>>> type(graphite) # doctest: +SKIP
<class 'javelin.structure.Structure'>
>>> PbTe = Structure(read('tests/data/PbTe.cif')) # doctest: +SKIP
>>> print(PbTe) # doctest: +SKIP
Structure(Pb4Te4, a=6.461, b=6.461, c=6.461, alpha=90.0, beta=90.0, gamma=90.0)
>>> type(PbTe) # doctest: +SKIP
<class 'javelin.structure.Structure'>
ASE can also be used to write file to many file-formats using
ase.io.write()
>>> from ase.io import write # doctest: +SKIP
>>> write('output.xyz', graphite.to_ase()) # doctest: +SKIP
>>> write('output.png', graphite.to_ase()) # doctest: +SKIP